Abstract
Background: Autologous chimeric antigen receptor T (CAR-T) cell therapies show significant clinical activity against hematologic malignancies including multiple myeloma (MM). Numerous factors influence the effectiveness of this therapeutic approach. The fitness of a CAR-T cell product is an important factor affecting T cell engraftment and persistence, a pre-requisite for effective CAR-T cell therapy. In an effort to deconstruct the cellular heterogeneity of CAR-T cell products and explore relationships between T cell states within the cellular product, gene expression and CAR-T cell engraftment following adoptive transfer, we performed single cell RNA-sequencing (scRNA-seq) on CAR-T cell products from a phase I trial of BCMA-specific CAR-T cells in relapsed/refractory MM patients that included cohorts receiving a similar CAR-T cell dose with and without preconditioning with cyclophosphamide (NCT02546167, Cohen et al, J Clin Invest 2019).
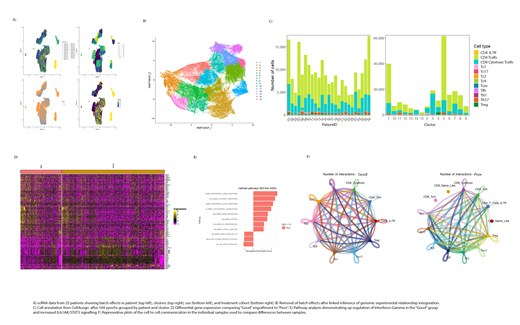
Method: scRNA-seq was performed on 25 unique products with an average of 8823 cells captured per product at an average depth of 40779 reads per cell. Batch effects were controlled by integration using established metrics for quality control. The linked inference of genomic experimental relationships (LIGER) method exhibited the most effective integration of 228,752 single cells across the 25 products. Annotation was completed by comparing transcriptomes of cells to the Blueprint/ENCODE references of pure immune cells. Differential gene expression (DGE) analysis was conducted by both Wilcoxon rank sum test testing and negative binomial distribution. Gene enrichment analysis was performed by comparing DGE with the Molecular Signatures Database v7.4. Cell to cell communication utilized the KEGG signaling pathway maps and curated lists of interactions from the literature. The intercellular communication probability was estimated on the DE genes before statistically significant intercellular communications were calculated by a permutation test, with dominant senders, receivers, mediators, and influencers identified using graph theory. Patients were grouped based upon "good" or "poor" engraftment using a peak blood vector copies per cell cutoff of 10,000. Association of transcriptional variation with neurotoxicity was explored for all patients.
Results: The CD4:CD8 ratio of 2.51 single cells in the data was comparable to the observed average CD4:CD8 ratio of total T cells in the product based upon flow cytometric analysis of the products at harvest. Cell annotation showed significant heterogeneity of CD4+ and CD8+ T cells with cells exhibiting a CD4 Tcm transcriptional profile comprising the largest subset of T cells within the product. DGE analysis found 205 genes that were up or down regulated between the "good" vs "poor" engraftment phenotypes with 75 genes being detected using both mathematical approaches. CAR-BCMA expression was increased in "good" engraftment groups which has not previously been shown in CAR sorted cells. Within both CD4+ and CD8+ T cell subsets, transcriptional pathways associated with the RXF5/RFXAP/RFXANK transcriptional activator complex and the IL-2/STAT5 signaling were identified as upregulated in the "good" engrafted products. Cell to cell communication analysis for both secreted signaling and cell to cell contact revealed similar ligand-receptor interaction differences between the engraftment groups. Within the neurotoxicity groups CAR-BCMA expression was not associated with either neurotoxicity or with high and low neurotoxicity effects in the product. High vs low neurotoxicity showed a shift with interferon gamma and cytokine-cytokine interactions. We also detected IL-6/JAK/STAT3 signaling activation increases in this group.
Conclusion: The RFX5/RFXAP/RFXANK pathway associated with MHC class II expression and IL-2/STAT5 signaling show a significant association with engraftment of BCMA-specific CAR-T cells. Although IL-2 signaling is well known to be critical to T cell survival and a potential key driver for long-term persistence, the role of the RFX5 transcriptional activator complex in T cells, outside of its important role in regulating MHC expression, is largely unknown. Both pathways deserve further investigation. Cell to cell communication between engraftment groups suggests CD4/CD8 interactions that might be beneficial to engraftment at the product manufacturing stage.
Garfall: Amgen: Honoraria; CRISPR Therapeutics: Research Funding; GlaxoSmithKline: Honoraria; Janssen: Honoraria, Research Funding; Novartis: Research Funding; Tmunity: Research Funding. Bu: Novartis: Current Employment, Patents & Royalties: Co-inventor on patent applications. Orlando: Novartis: Current Employment. Brogdon: Novartis Institutes for Biomedical Research: Current Employment. Pruteanu-Malinici: Novartis: Current Employment. Cohen: Janssen: Consultancy; Oncopeptides: Consultancy; Genentech/Roche: Consultancy; BMS/Celgene: Consultancy; AstraZeneca: Consultancy; Novartis: Research Funding; Takeda: Consultancy; GlaxoSmithKline: Consultancy, Research Funding.